Contents
- Prediction of preterm birth
- Quantification of joint damage in Rheumatoid Arthritis
- Prediction of COVID-19 severity
- An annotated brain tissue dataset
- Identification of tiger mosquitos through citizen science
- XAI-AGE: an explainable predictor of chronological age
- HunCRC: an annotated large-scale digital pathology dataset in colorectal cancer
- Nightingale Open Science challenge - Predicting High Risk Breast Cancer
- Related publications
Our research group has a long-standing interest in the application of artificial intelligence (AI) in various scientific disciplines. We have been involved in several projects that aim to develop AI-based tools for the analysis of biological data, let that be medical images, omics data, or other types of large-scale datasets. Below, we provide a brief overview of some of our projects in this area.
Prediction of preterm birth
Preterm birth is a major public health concern, as it is the leading cause of newborn deaths worldwide. Thus, longitudinal whole-blood transcriptomic data at exon-level resolution and plasma proteomic data on 216 women were collected and a crowdsourcing DREAM (Dialogue for Reverse Engineering Assessments and Methods) challenge was initiated for better prediction of gastational age and preterm birth.
Our group participated in this project and developed the top ranking machine learning model of the challenge.
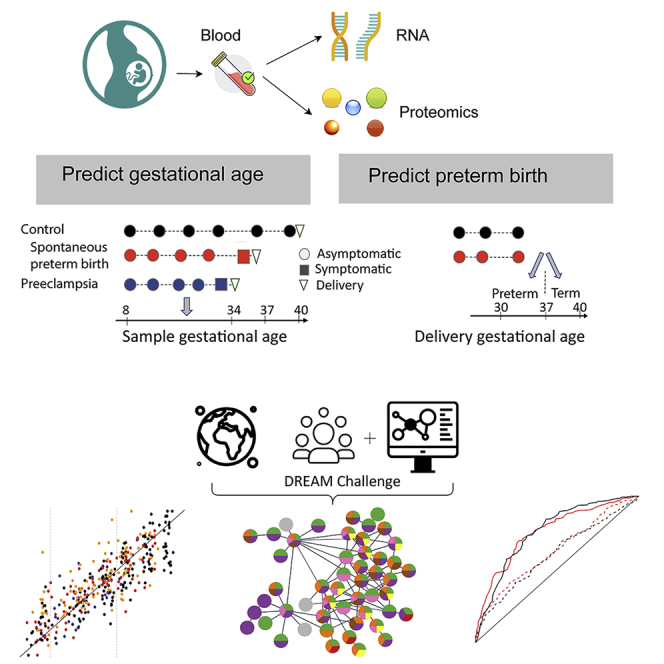
The combined results of the best performing models submitted by the participants indicate that whole-blood gene expression predicts ultrasound-based gestational ages in normal and complicated pregnancies and, using data collected before 37 weeks of gestation, also predicts the delivery date in both normal pregnancies and those with spontaneous preterm birth. Based on samples collected before 33 weeks in asymptomatic women, the analysis suggests that expression changes preceding preterm prelabor rupture of the membranes are consistent across time points and cohorts and involve leukocyte-mediated immunity. Models built from plasma proteomic data predict spontaneous preterm delivery with intact membranes with higher accuracy and earlier in pregnancy than transcriptomic models.
Quantification of joint damage in Rheumatoid Arthritis
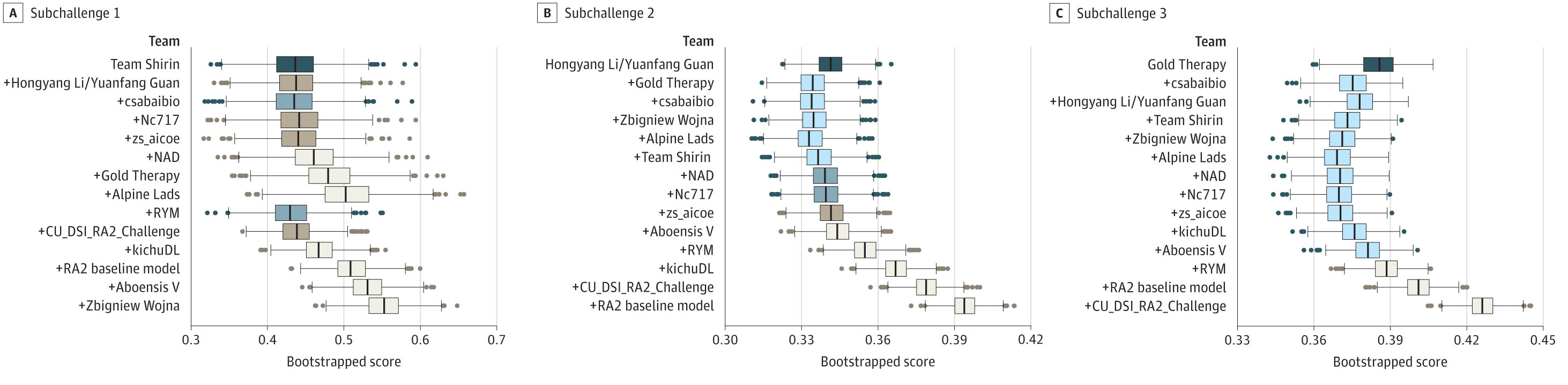
Rheumatoid arthritis (RA) is a chronic autoimmune disease that affects the joints. It is characterized by inflammation of the synovial membrane, which leads to joint damage and loss of function. The severity of joint damage is typically assessed using radiographic images, and the most widely used scoring system is the Sharp/van der Heijde score. This score is based on the evaluation of hand and foot radiographs, and it is used to monitor the progression of joint damage in RA patients. However, the scoring process is time-consuming and requires a high level of expertise. Therefore, in the framework of a DREAM challange, we have developed an AI-based tool for the automatic quantification of joint damage in RA patients. The tool uses deep learning to analyze radiographic images and to provide a quantitative measure of joint damage.
The model of our group was among the top performing models in all of the three sub-challenges of the DREAM challenge.
Prediction of COVID-19 severity
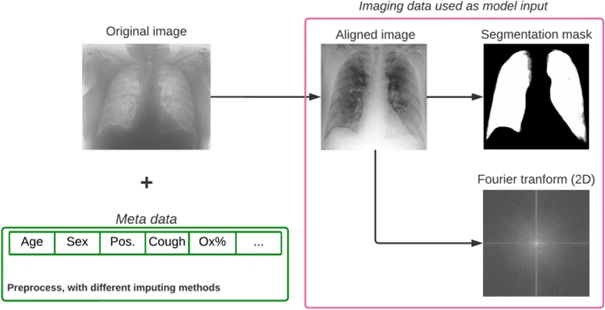
As an implementation of AI-based methods in COVID-19-related research, we took part in the "Covid CXR Hackathon—Artificial Intelligence for Covid-19 Prognosis" project, initiated in February, 2022 due to the significant challenges faced by Northern Italy during the early stages of the pandemic. The goal of the challenge was to develop an AI-based model capable of predicting the severity of COVID-19 upon admission based on chest X-ray (CXR) images and clinical metadata.
Through extensive analysis, we found that while CXRs provide valuable insights, they do not significantly enhance predictive power beyond the information available from other clinical data at admission.
This work highlights the potential of AI in improving medical prognosis by effectively combining different types of healthcare data.
An annotated brain tissue dataset
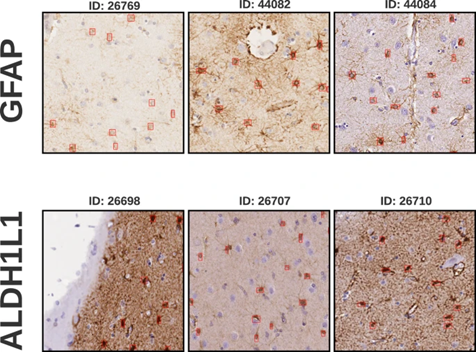
We have created and published an annotated brain tissue dataset for the training and evaluation of AI-based tools for the analysis of brain tissue images. Astrocytes, a type of glial cell, significantly influence neuronal function, with variations in morphology and density linked to neurological disorders. Traditional methods for their accurate detection and density measurement are laborious and unsuited for large-scale operations. We introduce a dataset from human brain tissues stained with aldehyde dehydrogenase 1 family member L1 (ALDH1L1) and glial fibrillary acidic protein (GFAP). The digital whole slide images of these tissues were partitioned into 8730 patches of 500x500 pixels, comprising 2323 ALDH1L1 and 4714 GFAP patches at a pixel size of 0.5019/pixel, furthermore 1382 ADHD1L1 and 311 GFAP patches at 0.3557/pixel.
Sourced from 16 slides and 8 patients our dataset promotes the development of tools for glial cell detection and quantification, offering insights into their density distribution in various brain areas, thereby broadening neuropathological study horizons.
These samples hold value for automating detection methods, including deep learning. Derived from human samples, our dataset provides a platform for exploring astrocyte functionality, potentially guiding new diagnostic and treatment strategies for neurological disorders.
Identification of tiger mosquitos through citizen science
Besides the above presented potential of crowdsourcing in developing classfication methods, its application in the field of public health is also promising. A recent approach in surveillance techniques is the use of smartphones and the Internet to enable novel community-based and digital observatories, where people can upload pictures of disease vectors (e.g. mosquitos) whenever they encounter them. This presents a great advantage over traditional surveillance methods, as these generally rely on catches, which requires regular manual inspection and reporting, and dedicated personnel, making large-scale monitoring difficult and expensive.
An example is the Mosquito Alert citizen science system, which includes a dedicated mobile phone app through which geotagged images are collected. This system provides a viable option for monitoring the spread of various mosquito species across the globe, although it is partly limited by the quality of the citizen scientists' photos. To make the system useful for public health agencies, and to give feedback to the volunteering citizens, the submitted images are inspected and labelled by entomology experts. Although citizen-based data collection can greatly broaden disease-vector monitoring scales, manual inspection of each image is not an easily scalable option in the long run, and the system could be improved through automation. Based on Mosquito Alert's curated database of expert-validated mosquito photos, we trained a deep learning model to find tiger mosquitoes (Aedes albopictus), a species that is responsible for spreading chikungunya, dengue, and Zika among other diseases.
The highly accurate 0.96 area under the receiver operating characteristic curve score promises not only a helpful pre-selector for the expert validation process but also an automated classifier giving quick feedback to the app participants, which may help to keep them motivated.
XAI-AGE: an explainable predictor of chronological age
Ageing is often characterised by progressive accumulation of damage, and it is one of the most important risk factors for chronic disease development. Epigenetic mechanisms including DNA methylation could functionally contribute to organismal aging, however the key functions and biological processes may govern ageing are still not understood. Although age predictors called epigenetic clocks can accurately estimate the biological age of an individual based on cellular DNA methylation, their models have limited ability to explain the prediction algorithm behind and underlying key biological processes controlling ageing. Our group has therefore developed XAI-AGE, a biologically informed, explainable deep neural network model for accurate biological age prediction across multiple tissue types.
We have shown that XAI-AGE outperforms the first-generation age predictors and achieves similar results to deep learning-based models, while opening up the possibility to infer biologically meaningful insights of the activity of pathways and other abstract biological processes directly from the model.
HunCRC: an annotated large-scale digital pathology dataset in colorectal cancer
Colorectal cancer (CRC) is one of the most common cancers worldwide, and as such, amoong the leading causes of cancer-related deaths. The prognosis of CRC patients is highly dependent on the stage of the disease at the time of diagnosis. Therefore, the accurate and early detection of CRC is of utmost importance. In the framework of the HUN-CRC project, we have created one of the first annotated large-scale digital pathology datasets for the training and evaluation of AI-based tools for the analysis of CRC tissue samples. It consist of 200 large-scale whole-slide images (approximately 500,000 x 500,000 pixels) with various pixel level segmentation masks on different anomalies in the biopsies. We pre-processed the dataset for downstream use and built machine learning baselines for others to later compare against.
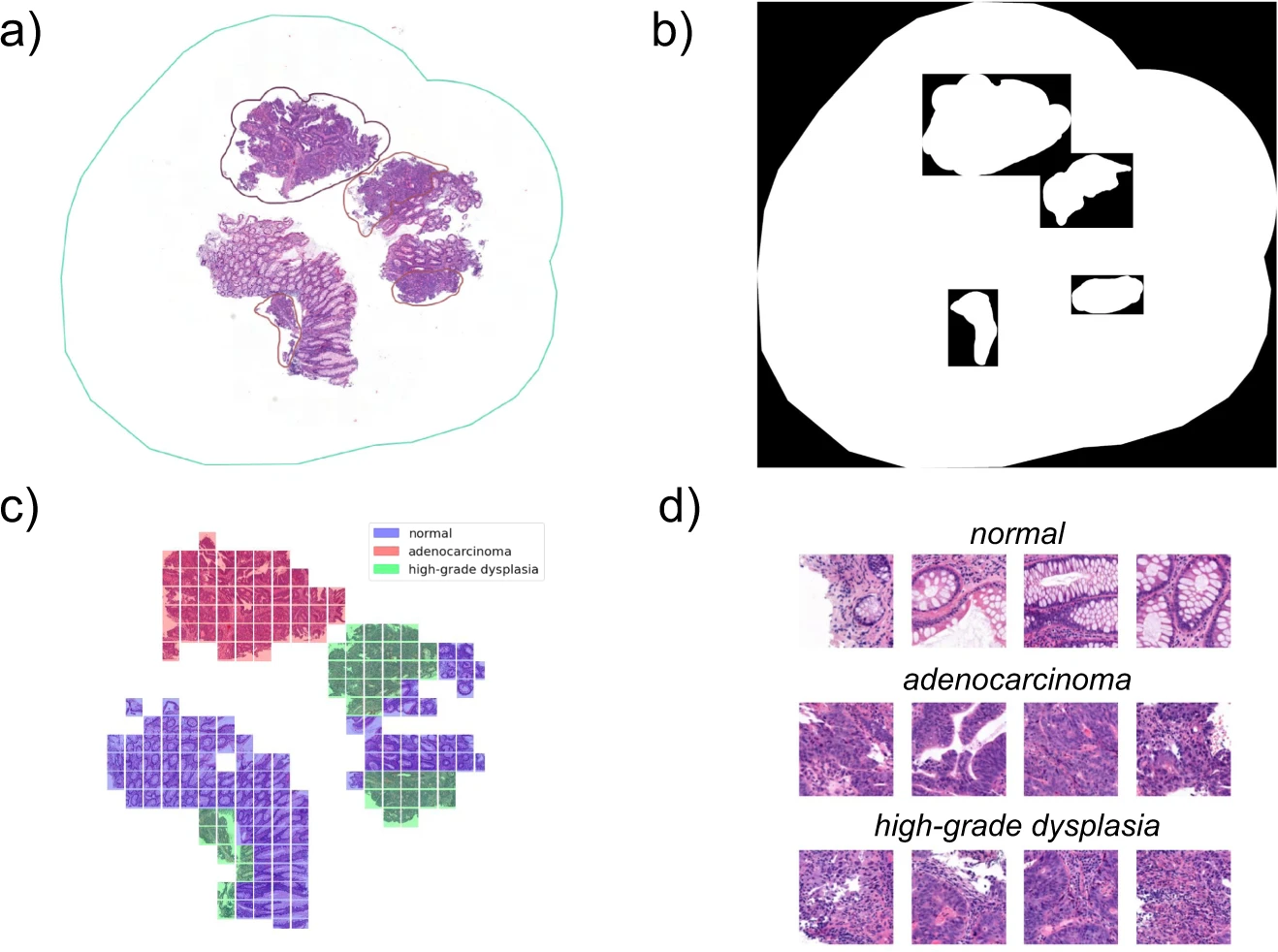
Nightingale Open Science challenge - Predicting High Risk Breast Cancer
Nightingale Open Science is a non-profit computing platform dedicated to advancing interdisciplinary medical research by providing access to a vast collection of deidentified medical data, including images and associated ground-truth labels, sourced globally from health systems. Launched in December 2021 with support from major funders, it aims to address the acute shortage of open datasets in medical research. The platform curates large-scale medical imaging datasets around unresolved medical challenges such as sudden cardiac death, cancer, and COVID-19, with a focus on leveraging machine learning to interpret complex imaging data. Hosted on their secure cloud infrastructure, these datasets are intended for non-profit research that could potentially uncover new patterns in disease and transform our understanding and treatment of these medical challenges.
The "Predicting High Risk Breast Cancer - Phase 1" challenge, launched by Providence St. Joseph, Nightingale OS, and The Association for Health Learning and Inference (AHLI), focused on analyzing digital pathology images to determine the stage of breast cancer from biopsy slides and aimed at fostering the development of algorithms to identify new patterns in these images. The dataset encompassed whole slide images (WSIs) from 4,335 breast biopsies of 3,425 patients from 2014 to 2020, with each biopsy's cancer stage as the primary label. This challenge called for creative solutions to overcome a range of obstacles. Faced with a vast dataset of terabytes, heterogeneity in patient slides, lack of annotations, and limited computational resources, we developed an efficient segmentation methodology for slide preprocessing, utilized a publicly available dataset with expert annotations for supervised pre-training of our feature extraction model and implemented a multiple instance learning (MIL) framework to accommodate variable input sizes and the absence of slide level labels.
Our method, reminiscent of a pathologist's visual slide analysis, secured state-of-the-art performance in the international challenge, earning first prize.
Demonstrating potential in breast cancer staging, our approach requires no external information or additional tests, offering both interpretability and explainability for a task previously deemed unattainable. The adoption of these techniques could significantly enhance breast cancer diagnosis and treatment, leading to improved patient outcomes.
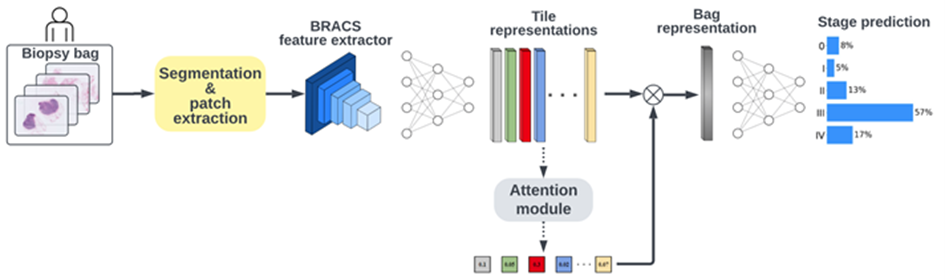
The "Predicting High Risk Breast Cancer - Phase 2" challenge built upon the initial phase, which concluded in January 2023, by concentrating on a more balanced subset of biopsy data. This phase aimed to represent various cancer stages and minority race and ethnicities more evenly, a shift from Phase 1's skew towards earlier cancer stages and lower minority representation. The challenge involved analyzing cancer stages from biopsy images of 1,000 biopsies, corresponding to 842 patients from 2014 to 2020, with an updated scoring method based on the area under the ROC-curve (AUC). The baseline score for this phase was established using CLAM, a weakly supervised learning method for classifying whole slide images, achieving a baseline AUC of 0.68. Our initial staging methodology, which was developed during the first phase of the contest, underwent significant refinements. In the second phase of the contest, we leveraged the vision transformer architecture and self-supervised pretraining. Our methodology, which built upon the state-of-the-art HIPT architecture enhanced by transfer learning, was capable of capturing spatial correlations at multiple resolutions through multi-step embedding techniques. It also eliminated the supervised bias of transfer learning and the inductive biases inherent in CNNs.
Our improved method led to our success in winning the second phase of the competition. These results had the potential to drive further research in automated staging using solely whole-slide imagery data, paving the way for new discoveries in this field.
Related publications
- Tarca et al. Crowdsourcing assessment of maternal blood multi-omics for predicting gestational age and preterm birth. Cell Rep Med. 2(6), 100323 (2021). DOI: 10.1016/j.xcrm.2021.100323
- Sun et al. A Crowdsourcing Approach to Develop Machine Learning Models to Quantify Radiographic Joint Damage in Rheumatoid Arthritis. JAMA Netw Open., 5(8), e2227423 (2022). DOI: 10.1001/jamanetworkopen.2022.27423
- Olar et al. Automated prediction of COVID-19 severity upon admission by chest X-ray images and clinical metadata aiming at accuracy and explainability. Sci Rep 13, 4226 (2023). DOI: 10.1038/s41598-023-30505-2
- Olar et al. Annotated dataset for training deep learning models to detect astrocytes in human brain tissue. Scientific Data 11 (1), 96 (2024). DOI: 10.1038/s41597-024-02908-x
- Pataki et al. Deep learning identification for citizen science surveillance of tiger mosquitoes. Sci Rep 11, 4718 (2021). DOI: 10.1038/s41598-021-83657-4
- Pataki et al. HunCRC: annotated pathological slides to enhance deep learning applications in colorectal cancer screening. Sci Data 9, 370 (2022). DOI: 10.1038/s41597-022-01450-y
- Prosz et al. Biologically informed deep learning for explainable epigenetic clocks. Sci Rep 14, 1306 (2024). DOI: 10.1038/s41598-023-50495-5
- Biricz and Bedőházi et al. Breast cancer staging with deep learning solely from histopathology whole slide images. npj Breast Cancer (2023). - under review
- Bedőházi et al. Breast Cancer Stage Prediction with Deep Learning. ML4H Conference invitation and poster presentation (2023).
